- Research
- Open access
- Published:
Integrated analysis, transcriptome-lipidome, reveals the effects of INO- level (INO2 and INO4) on lipid metabolism in yeast
BMC Systems Biology volume 7, Article number: S7 (2013)
Abstract
Background
In the yeast Saccharomyces cerevisiae, genes containing UASINO sequences are regulated by the Ino2/Ino4 and Opi1 transcription factors, and this regulation controls lipid biosynthesis. The expression level of INO2 and INO4 genes (INO-level) at different nutrient limited conditions might lead to various responses in yeast lipid metabolism.
Methods
In this study, we undertook a global study on how INO-levels (transcription level of INO2 and INO4) affect lipid metabolism in yeast and we also studied the effects of single and double deletions of the two INO-genes (deficient effect). Using 2 types of nutrient limitations (carbon and nitrogen) in chemostat cultures operated at a fixed specific growth rate of 0.1 h-1 and strains having different INO-level, we were able to see the effect on expression level of the genes involved in lipid biosynthesis and the fluxes towards the different lipid components. Through combined measurements of the transcriptome, metabolome, and lipidome it was possible to obtain a large dataset that could be used to identify how the INO-level controls lipid metabolism and also establish correlations between the different components.
Results
In this study, we undertook a global study on how INO-levels (transcription level of INO2 and INO4) affect lipid metabolism in yeast and we also studied the effects of single and double deletions of the two INO-genes (deficient effect). Using 2 types of nutrient limitations (carbon and nitrogen) in chemostat cultures operated at a fixed specific growth rate of 0.1 h-1 and strains having different INO-level, we were able to see the effect on expression level of the genes involved in lipid biosynthesis and the fluxes towards the different lipid components. Through combined measurements of the transcriptome, metabolome, and lipidome it was possible to obtain a large dataset that could be used to identify how the INO-level controls lipid metabolism and also establish correlations between the different components.
Conclusions
Our analysis showed the strength of using a combination of transcriptome and lipidome analysis to illustrate the effect of INO-levels on phospholipid metabolism and based on our analysis we established a global regulatory map.
Background
Phospholipid synthesis in the yeast Saccharomyces cerevisiae is a complex process that involves regulation by both genetic and biochemical mechanisms [1–3]. The activity levels of phospholipid synthesis enzymes are controlled by gene expression, e.g., transcription, and by other factors, i.e., lipids, water-soluble phospholipid precursors and products, and covalent modification by phosphorylation.
At the transcription level, the heterodimeric Ino2/Ino4 activator and the transcription factor Opi1 are global regulators affecting the expression of a large number of phospholipid biosynthetic genes [4–6]. Opi1, containing a leucine zipper motif, has been known as a negative regulator of phospholipid biosynthesis and it can also repress the transcription of INO2 and INO4 [7, 8]. In the absence of Opi1, the transcriptional level of both INO2 and INO4 (so called "INO -level") will be up-regulated compared to the reference strain. Ino2p and Ino4p binds to so-called inositol-choline response elements (ICRE), and ICRE-bound Ino2p can interact with coactivator complexes such as Snf1 kinase that has histone kinase function, the SAGA complex, and the TFIIB complex when OPI1 is disrupted or not presented in the nucleus [9, 10]. Opi1 is also necessary for repression of ICRE-dependent transcription when inositol-choline is present in excess [8, 11]. Under this condition, Opi1 is localized in the nucleus [12] and prevents Ino2 from activation of target genes by recruiting the pleiotropic co-repressors such as the Cyc8/Tup1 complex [13, 14] or Sin3p [15–19]. Consequently, Ino2 variants (which are defective in interacting with Opi1) lead to the repression of phospholipid synthetic genes [20]. However, a genome wide transcription analysis of the effect of varying INO-levels that effect lipid metabolism has never been studied before.
It is known that the transcription of the phospholipid biosynthetic genes is maximally repressed in the presence of the phospholipid precursors inositol and choline (IC) [4, 21, 22]. A highly conserved 10bp-element (5'-CATGTGAAAT-3'), in at least one copy is found in the promoters of the lipid co-regulated genes such as INO1, CHO1, CHO2, OPI3, FAS1, FAS2, ACS2, and ACC1 [23–28]. This element has been shown to be both necessary and sufficient for the IC response, the so called "inositol/choline-responsive element" or "inositol-sensitive upstream activating sequence" (ICRE or UASINO motifs)[23, 24, 29]. These motifs are bound by a heterodimer of positive regulators Ino2p and Ino4p containing a basic helix-loop-helix (bHLH) structural motif [21, 30–32] which is necessary and sufficient for dimer formation and specific interaction with the UASINO motif [10, 32, 33]. Recent studies have shown that expression of several genes, probably unrelated to phospholipid metabolism, is also affected by Ino2p and Ino4p [22, 29, 34]. Importantly, over-expression of INO2 (but not of INO4) counteracts IC repression, suggesting Ino2p as a possible target of the signal transduction pathway triggering IC repression [35, 36]
At the biochemical mechanism level, the level of phosphatidic acid (PA) is controlled by the biochemical regulation of key phospholipid synthesis enzymes [12] and it plays a central role in the regulation of phospholipid synthesis gene expression [1, 33, 37].
In this study, we undertook a global study of lipid metabolism in response to different INO-levels (INO2 and INO4): (1) normal INO-level using wild-type (CEN.PK113-7D), (2) high INO-level using an opi1 Δ strain, and (3) low (or rather absent) INO-level using an ino2 Δ ino4 Δ double deletion strain. Moreover, we also focused on the deficient factors as a sub story comparing the individual knockout strains (ino2 Δ and ino4 Δ) with the double deletion strain. Using a systems biology approach [38] a global regulatory model for lipid metabolism could be established. With 2 types of nutrient limitations (carbon and nitrogen) and different INO-level, we were able to see the effect on expression level of the genes involved in lipid biosynthesis and the fluxes towards the different lipid components. Through combined measurements of the transcriptome, metabolome, and lipidome it was possible to obtain a large dataset that could be useful to identify the effect of INO-level and also establish correlations between the different components.
Methods
Materials
All chemicals were reagent grade. Phospholipids, fatty acid methyl ester and neutral lipids standards were purchased from Sigma.
Agar spot test on SD media with different inositol concentrations
The reference strain CEN.PK 113-7D and 5 mutants as shown in table 1 were grown on SD agar plate (containing Yeast Nitrogen Base without amino acids and inositol, Formedium LTD, England) for 48 hours. The SD agar plates were supplemented with 0, 1.39, 75 or 220 µM of inositol.
Strains, cultivation, and fermentation profile
The S. cerevisiae strains used in this study were a prototrophic strain CEN.PK 113-7D (Mata Mal2-8c SUC2) [39] and its derivative (opi1∆, ino2∆, ino4∆, and ino2∆ino4∆) supplied by Peter Kötter (Frankfurt, Germany). All strains in this study were prototrophic and with mating type a (Table 1). Steady-state aerobic chemostat cultures were grown at 30 ˚C in 1.2 L bioreactors (DASGIP, Germany) with working volume of 0.5 L using a dilution rate of 0.10 (±0.005) h-1. For the C-limited and N-limited cultures, the medium composition was the same as in a previous study [40, 41] which contained 75 μM of inositol and 1mM of choline [24, 42]. The pH was controlled at 5.00 ± 0.05 with 2M KOH and dissolved oxygen was kept above 30%. Chemostat cultivation ensured that metabolic and regulatory changes observed were specific to the INO-level and also the disruptions of INO2 and/or INO4, and not complicated by external effects resulting from different specific growth rates.
Samples were harvested from the cultivation media every second hour and immediately filtered through a 0.45 μm pore-size cellulose acetate filter (VWR) and stored at -20 ˚C until analysis. Biomass production was evaluated by measuring of optical density (OD600) and dry cell weight. Glucose, glycerol, ethanol, and acetate concentrations were determined by HPLC analysis using an Aminex HPX-87H column (Biorad, Hercules, CA) [43].
Transcriptome analysis
Transcriptome data acquisition
Samples for RNA extraction were taken after 50 h (i.e. 5 retention times) of steady-state by rapidly taking 20 ml of culture and mixing with 30 ml of crushed ice in a 50 ml Falcon tube to cool down the samples immediately. The cells were harvested by centrifuging at 4000 rpm and 2˚C for 3 min, and then frozen in liquid nitrogen and stored at -80˚C until subsequent RNA extraction. The cells were mechanically disrupted using FastPrep homogenizer (MP Biomedicals) and total RNA was isolated using the RNeasy Mini Kit (QIAGEN). The quality of total RNA was assessed using an Agilent 2100 Bioanalyzer (Agilent Technologies) with RNA 6000 Nano LabChip kit (Agilent Technologies). The labeled RNA was synthesized using the GeneChip 3' IVT Express Kit (Affymetrix), which was then hybridized onto the GeneChip Yeast Genome 2.0 Arrays (Affymetrix). Staining and washing of the hybridized arrays were carried out on the GeneChip® Fluidics Station 450 (Affymetrix) and scanned using the GeneChip Scanner 300 7G (Affymetrix). All transcriptome data of this study can be found at Gene Expression Omnibus with accession number GSE36298.
Transcriptome data analysis
The transcriptome data were analyzed using Bioconductor in R. Raw data were normalized and processed together with Probe Logarithmic Intensity Error (PLIER http://media.affymetrix.com/support/technical/technotes/plier_technote.pdf). Pairwise T-test analysis was performed to determine the genes whose expression level is significantly changed due to INO-level, as well as the sufficiency factor. The calculated P-values were corrected for multiple testing by FDR method. A cut-off value of adjusted P value<0.01 was set to assess statistical significance.
Lipid data acquisition
Total lipid extraction
The lipid extraction method was adapted from Bligh and Dyer[44]. First, 15 mg of freeze-dried cell pellets were treated with 1 unit μl-1 of zymolyase digesting buffer (1.2 M glycerol, 100 mM sodium thioglycolate, 50 mM Tris-sulfate, pH 7.5) at 37°C for 15 min, followed by centrifugation at 3000 rpm for 3 min to collect the spheroplast, which was mixed with internal standards (heptadecanoic acid and glyceryl tri-heptadecanoate, 25 µg of each). After the addition of 7 ml of chloroform-methanol (2:1, v/v), the mixture was shaken horizontally at 300 rpm 4°C for 3 h, mixed with 1.7 ml of sodium chloride solution (0.73%) and centrifuged at 3000 rpm 4°C for 4 min for phase separation. The lower (organic)-phase was collected and the remaining was re-extracted with 5 ml of chloroform-methanol (85:15 v/v). The lower (organic)-phase was collected and pooled with the previous organic fraction and kept at -20°C until further analysis.
Lipid class separation, identification, and quantification using HPLC-CAD
Lipid separation and quantification were performed using our developed method [45]. Lipid separation was accomplished by HPLC (Dionex) equipped with charge aerosol detector; CAD (Corona) and the gas connected was nitrogen gas with 35 psi gas pressure. All the separated fractions were then collected by automated fraction collector; AFC-3000 (Dionex). A 20 μl volume of sample was injected in to the Luna 5 µm HILIC 200 Å 100 × 3.0 mm LC Column (Phenomenex). The flow-rate was 0.8 ml/min and the column temperature was kept at 25˚C during all runs. The chromatogram was record at 10 Hz frequency and gain for 100 pA. The polar and neutral lipid classes were separated by three solvent mixtures and gradient systems as follow: (A) hexane-acetic acid (99:1, v/v); (B) acetone-isopropanol-acetic acid (29:70:1, v/v/v); (C) water-acetone-isopropanol-acetic acid (9:20:70:1, v/v/v/v). Triethylamine (0.08%, v/v) was added to the solvent C to adjust pH. The samples were injected at time 0 and the gradient profile started at 100% of solvent A and the solvent B was gradually increased to 5% in 14 min and it was always kept at 5% along the process. At 15 min time point, solvent C was slowly entering to the system and rising up to 40% in 5 min. Then solvent C was slowly increased until 45% in 20 min. Finally, the gradient was reduced from 5% to 0% of solvent B and from 45% to 0% of solvent C in 5 min and then maintained at 100% of solvent A for 5 min. In total, the solvent program for the separation of all lipid classes took 45 min.
Identification and quantification
Pure lipid standards were analyzed individually using chromatography to confirm their retention times and purity. Lipid standards were also co-eluted together with samples to identify peaks in unknown samples. Solutions of known concentrations of different lipid classes were mixed and lipid standard curves were generated to study the linearity of the detection method and to quantify lipid classes in unknown samples. Calibration curves were prepared for 5-1000 μg ml-1 of PA, PE, PC, PS, PI, ES, TAG, FA, and ES. Each concentration of the standard solutions was injected twice and the average log10 peak area for each lipid was plotted against the absolute amount of lipid. Correlation (r2) was determined for all curves by linear regression.
Fatty acid methylesters (FAMEs) analysis
To quantify the distribution of fatty acid long chain species, we used standard procedure developed in our laboratory which is based on the previous protocol by Khoomrung et al. [46]. Briefly, 10 mg of freeze-dried samples was mixed with 4 mL of hexane, 2 mL of 14% BF3 (in Methanol) and 5 µg of internal standard (17:0) fatty acid standard was added. The sample was then flushed into the tube's head space with nitrogen gas for 30s and closed tightly with a Teflon screw cap. The tube was placed in a vessel containing 30 mL of milliQ water and then sealed with TFM screw cap. The tube was heated using microwave digestion system (milestone start D, Sorisole Bergamo, Italy) equipped with rotor PRO-24. The temperature programming of microwave digestion was ramped (from room temperature) to 120 ˚C within 6 min and maintained for 10 min. After cooling down sample at the room temperature, 2 mL of milliQ water was added and shaken vigorously for 1 min and centrifuged at 2500 rpm for 5 min. The upper phase (hexane phase which contained the FAMEs) was analysed by GC-MS.
The FAMEs were separated and quantified using Focus GC ISQ single quardrupole GC-MS (Thermo Fisher scientific, Germany). The separation of FAMEs was performed on Zebron (ZB-WAX) GC column (30 m × 0.25 mm I. D., 0.25 μm film thickness) from Phenomenex, Macclesfield, UK. Sample was injected in splitless injection mode (1µL at 240 ˚C) and Helium was a carrier gas (1 mL/min). The column temperature was initially set at 50 °C (1.5 min), then temperature was ramped to 180 °C (25°C/min) for 1 min, then increased to 220 °C (10°C/min) and held for 1 min. Finally, temperature was increased to 250 °C (15°C/min) and held for 3.0 min. Mass transfer line and ion source were set at 250 °C and 200 °C, respectively. The FAMEs were detected with electron ionization (70 eV) in scan mode (50-650 m/z) and selected ion monitoring mode at m/z 55, 67, 74 and 79 (for quantitative analysis). The identification of unknown FAMEs was achieved by comparing their retention times and mass spectrum profiles with known standards (Sigma-Aldrich, USA). The quantification of FAMEs was performed using QuanBrowser function in Xcalibur software version 2.0 (Thermo Fisher Scientific). According to the serial dilution of FAME mix standards and were normalized according to the internal standard fatty acid C17:0. The average molecular weights of each PL (Table.S1 in additional file 1) were used for mg/gDW and mmol/gDW units conversion (were later used for metabolic fluxes analysis).
Integrated analysis
The statistical adjusted P-values of each hypothesis testing were overlaid on the three networks graph of Gene Ontology, Transcription factor-gene interaction and genome-scale metabolic model iIN800 [47] (metabolite-gene interaction). Then reporter algorithm [48] was performed to obtain significant values (reporter p-value) of GO terms, Transcription factors (TF) and metabolites. The Platform for integrative analysis of omics data (PIANO) package for R ([49]available at http://www.bioconductor.org/packages/2.13/bioc/html/piano.html) has been performed for integrated analysis. All the features presented in heatmaps are those features that have reporter P-value < 0.001 and P-value < 0.01 were considered for INO-level and deficient factors, respectively.
Results
Inositol is essential in the low INO-level yeast
To estimate the required inositol concentration for low INO-level (either ino2Δ, or ino4Δ, or double deletion of them), a spot test with the 5 yeast strains on different concentrations of inositol were performed (Figure 1A). Since the Ino2-Ino4 heterodimer regulates positively the expression of INO1 (a structural gene for inositol-1-phosphate synthase which is required for inositol synthesis) and other phospholipid genes containing UASINO element [31, 50], deletion of INO2 and INO4 results in a requirement for supplementation of myo-inositol. However, myo-inositol is included in our minimal medium in a concentration of 250 µg/L (corresponding to about 1.39 µM) [41]. The spot test showed that opi1Δ as well as reference strain are prototroph for inositol but at least 75 µM of inositol is sufficient for low INO-level strains (either single or double deletion of INO2 and INO4) to grow. However, higher concentrations of inositol (even 200 µM) did not make much difference in term of growth. Based on this we conclude that the low INO-level strains are auxotroph for inositol, and this leads us to an experimental design using 75 µM of inositol in the medium for all the chemostat cultures.
Procedures of integrated analysis to reveal the effects of INO- level ( INO2 and INO4 ) on lipid metabolism in yeast. Spot test of all strains on SD agar plates with different inositol concentrations (A). Off gas CO2 profile of each mutant strain during the chemostat cultivation at C-limitation and N-limitation (B). Schematic analysis of a workflow for integrated analysis, transcriptome-lipidome, of INO-level (key factor) and deficient factor effect on lipid biosynthesis (C).
Cell physiology and carbon metabolism
To see the effects on cell growth of each mutant strain, 5 strains (the reference, opi1Δ, ino2Δino4, ino2Δ, ino4Δ) were grown in batch cultivations using a defined minimum medium and switched to chemostat immediately after glucose was depleted. Table 2 summarizes the basic physiological parameters for growth on glucose for all the strains in either C-limited or N-limited conditions. During the batch, ino2 Δ ino4 Δ double deletion had the highest maximum specific growth rate in the C-limited medium but the lowest maximum specific growth rate in the N-limited medium (Figure 1B). However, it was clear that the double deletion strain had longer lag phase during batch cultivation in both C-limited and N-limited conditions (25h and 60h, respectively). Interestingly, during the chemostat cultivation we found this double deletion strain had the lowest biomass yields compared to the other strains. Interestingly, we found that the opi1Δ strain exhibited metabolic oscillations in the glucose limited chemostat cultures. We therefore took the samples for transcriptome analysis at two time points, at the maximum and minimum level of CO2 production.
These two samples represent reductive-building (R/B) and reductive charging (R/C) of the yeast metabolic cycle respectively [51] and the transcriptional data analysis for both of them were performed to see the differences from 2 metabolic phases (Fig. S1 in additional file 1). The heatmap of reporter shows that the differences between these 2 metabolic phases mainly affects genes associated with differences in cell cycles period. According to the respirative quotient (RQ) from Table 2, the phase of the population at maximum CO2 production showed about 20% higher RQ compared with all the other strains at C-limitation. Based on this we therefore decided to exclude this sample and only use the sample from the minimum CO2 production as a high INO-level representative at C-limitation condition.
Double deletion of Ino2 and Ino4 resulted in a substantial reduction (about 35% in C-limited and 55% in N-limited conditions) in biomass yield compared with the reference strain (Table 2). Interestingly, deletion of Opi1 resulted in a small reduction (approx. 25%) in biomass only at N-limited but not at C-limited conditions.
INO-level in nutrient-limited conditions
The levels of INO-gene (INO2 and INO4) expression in both C-limited and N-limited conditions were investigated according to the expression level from micro array results (Figure 2). Even though there were metabolic cycle patterns with the opi1 Δ strain grown at C-limited condition, the expressions of the INO genes were consistently high (Figure 2A). This evidence shows that INO genes are up-regulated when their repressor, Opi1, is absent. At C-limited condition, INO levels are low in both the two single and the double deletion strains (Figure 2A and 2C). Interestingly, the INO4 gene seems to be expressed at a high level at N-limitation (Figure 2B) and it possibly also tries to compensate the failure of INO2 expression by up-regulating its expression in the ino2 Δ strain at N-limitation (Figure 2D).
Global transcriptome changes due to INO-level in nutrient-limited conditions
We used the Affymetrix DNA microarray platform to measure the expression level of all genes and access the global effect caused by the INO-level under nutrient-limited conditions (C-limited and N-lim). The transcriptome data were decomposed using principal component analysis (PCA) and Student's T-test analysis (α = 0.001). The transcriptome data are presented in Venn diagrams at C-limited (Figure 3A) and at N-limited (Figure 3B) showing that the high INO-level (opi1 Δ) strain had more genes being significantly changed at C-limitation. On the other hand, the low INO-level (ino2Δino4Δ double deletion) strain had more genes being significantly changed at N-limitation.
Transcriptional data analysis of 3 different INO -level strain. Venn's diagram of significant genes of mutant strains caused by different INO-level when focus on C-limitation (A) and N-limitation (B) separately, p-value < 0.001 were considered. (C) A heat map of overrepresented GO terms (Biological Process) of each factor comparison showing in the range of -4 to 4 of log(p-value). The green color indicates down-regulation and red indicates up-regulation compared to the reference strain.
From clustering of reporter GO terms (biological process at P < 0.001), 4 main clusters were identified (Figure 3C). Cluster 1 which is the largest group contains the genes involving the phospholipid biosynthesis, myo-inositol biosynthesis and transport, fatty acid metabolic process, cell conjugation, and ribonucleoside biosynthetic process which were highly up-regulated due to the deletion of OPI1 (high INO-level), especially at N-limited condition, but down-regulated at low INO-level at both C- and N-limitation. Cluster 2 contains genes involving maltose and sucrose catabolic process which were highly up-regulated due to the C-limitations. Cluster 3 contains genes involving nitrogen compound metabolism, amino acid and peptide transport, and proton transport which were down-regulated in the double mutant at N-limited condition. Cluster 4 contains genes involving amino acid biosynthesis, mitochondria biogenesis, and endoplasmic reticulum associated unfolded protein responses (ER-UPR) which were highly significant in the double mutant strain. These genes were up-regulated at low INO-level and N-limitations but down-regulated at C-limitation.
To identify specific transcriptional regulation of metabolism in response to deletion of INO2 and/or INO4, we performed transcriptome comparison of the three INO deficient mutants (defined as deficient factor in Figure 1C). For each strain we identified genes with significantly changed expression compared with the reference strain and then presented the results for the three strain as Venn diagrams for both C-limited (Figure 4A) and N-limited (Figure 4B) conditions. Even though it has been known that Ino2 and Ino4 form a heterodimeric transcription factor, we found some specific changes upon deletion of each of these genes (Figure 4A and 4C). Interestingly, ino4 Δ has more significant genes due to the absent of INO4 compared with the two other mutants at C-limitation. At N-limitation, on the other hand, the effect is significantly higher when both INO2 and INO4 were deleted. Interestingly, most of the genes involving nitrogen compounds metabolism and carbon utilization (Figure 4C, Cluster 1) were down-regulated due to the absence of INO2 and INO4 but up-regulated when only INO2 or INO4 were knocked out individually at N-limitation. Moreover, we found that the genes involving ribosome biogenesis and rRNA processing (in cluster 2 Figure 4C) are dramatically down-regulated when INO4 was knocked out especially at C-limited conditions. The evidence showed that Ino4 (probably together with other TFs) could possibly play role in ribosome biogenesis (which supports protein synthesis). Surprisingly, most of the genes involving protein translation (Cluster 3 in Figure 4C, including mitochondria translation) were up-regulated in the double mutant at N-limited conditions while they were only up-regulated when either INO2 or INO4 were deleted at C-limitation. Consistently, the genes involving phospholipid, inositol, and fatty acids (UASINO-contained genes in cluster 6, Figure 4C) were down regulated in all three strains at both C-limited and N-limited conditions. However, they were 2 other clusters that also captured down-regulated genes in all three INO-deleted strains, but different for the two nutrient limitations. The first group listed in cluster 4 (involving fatty acids biosynthesis and β-oxidation, TCA cycle, ATP, NAD, magnesium ion, and proton transport) were mainly down-regulated at N-limited condition. Cluster 5, on the other hand, contains genes involving ER-UPR and stress response, ethanol and amino acids biosynthesis that were mainly down-regulated only at C-limitation.
Transcriptional data analysis of 3 different INO -deficient strain. Transcriptional data analysis; Venn's diagram of significant genes of mutant strains caused by different deficient factors (single and double deletions) when focus on C-limitation (A) and N-limitation (B) separately, p-value < 0.01 were considered. (C) A heat map of overrepresented GO terms (Biological Process) of each factor comparison showing in range of -4 to 4 of log(p-value). The green color indicates down-regulation and red indicates up-regulation compared to the reference strain.
Effects of INO-level on fatty acid biosynthesis
With UASINO sites in their promoters, the expressions of fatty acid synthase genes such as ACC1, FAS1, and FAS2 are subject to control by the Ino2/Ino4 and Opi1 transcription factors [26, 52, 53]. At low INO-level strain, the fatty acid synthase genes (FAS1 and FAS2) were down-regulated at both C-limited and N-limited condition (Fig. S8 in additional file 1). However, single deletion of each INO gene seems to have effect only at C-limitation when FAS1, FAS2, and also ACC1 were down-regulated (Fig. S8 in additional file 1). In contrast, the high INO-level strains especially at N-limited condition showed an increase in FAS2 expression and none of the fatty acid synthase genes were down-regulated. This enhanced the ability to produce more FA needed for phospholipid biosynthesis. From the reporter metabolite heatmap (Fig. S2 in additional file 1), the FA pools and also fatty acyl-ACP were increasing for high INO-level strains especially at N-limited condition. All the low INO-level strains showed a dramatic decrease in FA and fatty acyl-ACP pools as reporter metabolites. However, it seems like the effects of lacking INO genes have higher effect at C-limited than at N-limited conditions (Fig. S2 in additional file 1). The transcription factors involving beta-oxidation, OAF1 and PIP2, were found as reporter TF which were highly up-regulated at high INO-level but down-regulated at low INO-level (Fig. S3 in additional file 1). Moreover, we found a significant increase in the free fatty acid (FA) pool of all the mutants compared to the reference strain (Figure 5). However, the low INO-level strains showed slightly higher amount of accumulated FAs due to the lower fluxes towards PLs. From the plots of correlation between each fatty acid chain and their elongase enzyme coding genes (Fig. S7 in additional file 1), we found a correlation between C18 fatty acid products (stearic and oleic acid) and the expression value of ELO1 to be about 0.65 while the correlation between C20 fatty acids and ELO2-ELO3 transcription were found in a negative direction (about -0.42 and -0.27, respectively). Presumably, C20 fatty acid might have a regulatory mechanism by repression their own elongase genes.
Fluxes through the different reactions of the lipid biosynthetic pathways. Fluxes through the different reactions of the lipid biosynthetic pathways for diferrent INO-levels (A) and diferrent deficient factors (B). All fluxes are shown in units of μmol/gDW/h. The normal font (left) and bold font (right) indicate the value from C-limitation and in N-limitation respectively. The level, in units of mg/gDW (±SD), of the different lipid species are shown in the table. (C = C-limited, N = N-limited).
Effects of INO-level on phospholipids biosynthesis and accumulation
As mentioned above the whole set of UASINO-contained genes were extremely up-regulated at high INO-level (Fig. S8 in additional file 1). However, to get a clearer picture about the carbon channeling in lipid metabolism at the metabolite level, the fluxes in lipid biosynthesis (in units of μmol/gDW/h) were calculated from the measured lipid profiles at all conditions (Figure 5) and this provides a clear picture of the changes in the flux distribution in response to the different factors evaluated. These fluxes were affected by many factors at several levels, such as gene transcription, protein phosphorylation, and enzyme activity. At high INO-level, the fluxes through CDP-DAG and phospholipids synthesis pathway were higher than the reference about 45-55% at C-limitation and N-limitation, respectively. Moreover, the expression of the genes coding for myo-inositol transporter (ITR1) and choline transporter (HNM1) were highly up-regulated in opi1 Δ. This supported the synthesis of PI and also enhanced the Kennedy pathway to produce more PC using choline and DAG as precursors and it was consistent with the highly up-regulation of the genes coding for Kenedy's pathway enzymes such as CPT1 and CKI1 (Fig. S8 in additional file 1). On the other hand, the ITR1 were constantly down-regulated in all low INO-level strains either double or single deletion in both limitation conditions. The low INO-level strain showed about 40-60% decreasing fluxes through the CDP-DAG pathway which caused a dramatically decrease in the phospholipid pool especially at N-limitation. The metabolite reporter heat map showed a large increase of PL pools in the high INO-level strain. In contrast, we found a dramatically decreasing of PLs in low INO-level compared to the reference strain.
Focusing on the deficient effects, double deletion of INO genes (especially at N-limitation) can cause more effect to the dramatically reduction of phospholipids than the effect of single deletions (Figure 5). About a 70% decrease in PLs can be found when both INO genes were deleted, but there were only 50% or 60% decreasing in PLs when INO2 or INO4 were deleted respectively. Even though this double deletion effects did not make much changes in the PLs pool at C-limitation, it still caused about 50% lower level of PI when compare to the single deletion strains. From the heatmap of reporter metabolites (Fig. S4 in additional file 1), S-adenosyl-L-methyonine or SAM (the only donor of PE methylation reaction) was strongly decreased in all low INO-level strains and this lead to a decrease of PLs (especially PC) since it is required for production of PC by Cho2 and Opi3.
Effects of INO-level on storage lipids biosynthesis and accumulation
The production rate and accumulation of PC can cause ER stress and UPR activation which lead to the up-regulation of TAG and ES biosynthesis [54]. At N-limitation, the low INO-level strain (double mutant) could produce and accumulate TAG and SE about 1.2 folds more when compared to the high INO-level strain (opi1 Δ) and about 1.5 folds when compare to the reference strain (Figure 5). Presumably, the down-regulation of LPP1 at both C- and N-limitation for the high INO-level strain led to a decrease in TAG biosynthesis and accumulation compared to the reference strain and other mutants (Fig. S8 in additional file 1). Consistently, almost the whole set of the ES genes (the green group in Fig. S8) were down-regulated compared to the reference and this was associated with a dramatically decreasing levels of ES and SE. This correlation between transcription of ES genes and ES level was also supported by the up-regulation of several ES genes in all low INO-level strains especially at N-limited condition (Fig. S8 in additional file 1). Interestingly, the double deletion strain showed a greater effect in increasing TAG only at N-limitation. This evidence was supported by the up-regulation of the ARE1 gene in the double deletion strain (Fig. S8 in additional file 1) which codes for the enzyme for the first step of SE biosynthesis. At C-limited condition, on the other hand, ino2 Δ ino4 Δ has a larger effect on decreasing the TAG level compared with the single deletion strains. Moreover, the fluxes through ES of the ino2 Δ strain was about 80% lower when compare to INO4 deletion and double deletion at C-limited condition.
Ino4 might have an extra function, beside the regulator of phospholipid biosynthesis, response to nitrogen starvation and amino acids starvations
It is possible that Ino4, but not Ino2, plays a role as a regulator for amino acid metabolism. It may not be Ino4 alone since Ino4, unlike Ino2, does not have a trans-activating domain (TAD) which is recognized for the RNA polymerase II complex [33, 55, 56]. In ino4 Δ, the genes involving ribosome biogenesis and assembly were extremely down regulated especially at C-limited conditions and this was exactly the responsive process for amino acids (and of cause nitrogen) starvation. There are indications that the induction of Ino4-regulated lipid biosynthesis genes may be connected to the immediate need of membrane material used for the autophagocytosis process [57]. This process is utilized by yeast in order to regulate the equilibrium between proteins and the diminishing set of amino acids due to starvation conditions [58]. From the reporter metabolite heatmap (Fig. S4 in additional file 1), most of the amino acids were found to be greatly down-regulated (especially at N-limitation) in all low INO-level strains which pointed an amino acid starvation response.
To identify the extra function of Ino4 on protein biosynthesis, we also performed the Pearson correlation analysis of INO4 to all other genes based on the normalized expression values from transcriptome data of all strains and all conditions. Using the cut off at 0.50 absolute correlation value, about 470 genes were selected as the high correlated genes of INO4 (91 genes were positively correlated and 380 genes were negatively correlated). The overrepresentation of gene ontology categories (biological process) were identified and analyzed using BiNGO, a Cytoscape plugin [59]. From the INO4 positively correlated genes listed in table S2 in additional file 1 it confirmed the main function of Ino4 (together with Ino2 as listed in table S4 in additional file 1) as a positive regulators of lipid biosynthesis and inositol-choline transport involved genes as it has been reported before by many researchers. However, the list of INO4 negatively correlated genes in table S3 (please see additional file 1) showed the new findings that Ino4 is actually play an extra role as the negative regulator of the translation process, protein biosynthesis and assembly, and also involves in mitochondrial translation. Ino2, on the other hand, had about 200 negatively correlated genes that passed through the cut off at 0.50 absolute Pearson correlation value. This particular group contains many interesting genes involving protein transport, protein catabolic process, and proteolysis which might also responds for amino acid starvation as summarized in table S5 (please see in additional file 1).
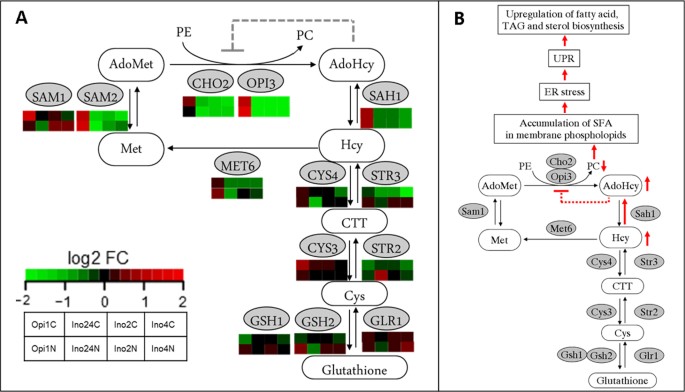
Linkage among sulfur-phospholipids, protein synthesis, and ER-UPR pathway
The double deletion of INO2 and INO4 at N-limitation showed some effects from amino acid starvation. UPR genes were also down-regulated when the synthesis level of amino acids was decreasing (less missed fold proteins). It also has been known that decreasing PC levels leads to the accumulation of saturated PC molecular species in the ER membrane which causes ER stress, UPR activation and these evidences lead to the up-regulation of FA, TAG, and sterol biosynthesis in the end [54]. Therefore, in the low INO-level strain, the KAR2 gene which is a responsive gene for ER-UPR was up-regulated especially at N-limitation. The high INO-level on the other hand, showed a large decrease in expression of KAR2 especially at C-limited condition. These evidences show that there is a linkage which plays a role in regulating the homeostasis among amino acids biosynthesis, phospholipids biosynthesis, and the ER-UPR pathway. As it has been reported before most of the genes coding for enzymes involved in cysteine-methionine biosynthesis, i.e. MET2, MET8, MET14, MET16, SAH1, SAH2, contain ICRE or UASINO sequences in their upstream regions [53], and these genes were up-regulated at high INO-level while OPI1 (the repressor) was disturbed. Consequently, we found down-regulation of MET6 and SAH1 and especially SAH2 in all low INO-level mutants, especially at C-limited condition. This leads to reduced synthesis and accumulation of storage lipids at C-limited condition compared to their references (Figure 6). This pointed out that amino acid synthesis genes might be controlled by the regulation of Snf1 kinase (via Gln3 or Gdh3) which is always activated at low glucose (C-limited) condition and consistent with our previous study [40]. Consistently, MET6 and SAH1 were up-regulated at high INO-level at N-limitations where Snf1 is consistently repressed by excess glucose in the culture media, and this leads to the slightly higher biosynthesis of TAG and hence TAG accumulation (Figure 6). Based on our findings and some supporting knowledge from the previous study [40], the overall regulation systems linking amino acids-protein synthesis, lipid metabolism and ER-UPR is summarised in Figure 7 which has kinase proteins and several transcription factors involved.
The coupled-reaction of methylations of phosphoethanolamine from S -adenosyl- L -methionine (AdoMet) by Cho2 and Opi3 enzymes. A comparison of expression level (log2 fold change) of each genes coding for sulfur-phospholipids coupled metabolism (A). The effects of low INO-level on ER stress and UPR inducing the up-regulation of FA and storage lipids (B).
Summary of the multilevel regulation network among amino acids biosynthesis, lipid metabolism, and ER-UPR. The figure illustrate the regulatory model of yeast metabolism involving protein and lipid metabolism showing all the effects from nutrients-responsive mechanism to the kinase, gene expression, and metabolite level.
Conclusions
Through integrated analysis of transcriptome and lipidome derived from robust experimental setup, it was possible to obtain a large-scale dataset that could be used to systematically identify correlations and associations between the different components. We were able to see the effect on expression of the genes involved in lipid biosynthesis and metabolic fluxes through lipid biosynthesis pathway. We found that there is an effect of Ino4, but not by Ino2, on the ribosome biogenesis and assembly which involves an amino acid starvation response. It points to an interesting link between lipid metabolism and the amino acid starvation response and we also found an effect of phospholipids on ER-UPR activation. Moreover, we found a close linkage among INO genes, amino acid genes, and probably Snf1 kinase in controlling lipid biosynthesis and accumulation. Following our analysis by genome-wide strategy and analysis of generated complex data by integrated analysis approach enable us to explore correlations and association of changes in a concerted fashion.
References
Carman GM, Han GS: Regulation of phospholipid synthesis in yeast. J Lipid Res. 2009, 50 (Suppl): S69-73.
Carman GM, Han GS: Regulation of phospholipid synthesis in the yeast Saccharomyces cerevisiae. Annu Rev Biochem. 2011, 80: 859-883. 10.1146/annurev-biochem-060409-092229.
Carman GM, Kersting MC: Phospholipid synthesis in yeast: regulation by phosphorylation. Biochem Cell Biol. 2004, 82 (1): 62-70. 10.1139/o03-064.
Santiago TC, Mamoun CB: Genome expression analysis in yeast reveals novel transcriptional regulation by inositol and choline and new regulatory functions for Opi1p, Ino2p, and Ino4p. J Biol Chem. 2003, 278 (40): 38723-38730. 10.1074/jbc.M303008200.
Ashburner BP, Lopes JM: Regulation of yeast phospholipid biosynthetic gene expression in response to inositol involves two superimposed mechanisms. Proc Natl Acad Sci USA. 1995, 92 (21): 9722-9726. 10.1073/pnas.92.21.9722.
Ashburner BP, Lopes JM: Autoregulated expression of the yeast INO2 and INO4 helix-loop-helix activator genes effects cooperative regulation on their target genes. Mol Cell Biol. 1995, 15 (3): 1709-1715.
White MJ, Hirsch JP, Henry SA: The OPI1 gene of Saccharomyces cerevisiae, a negative regulator of phospholipid biosynthesis, encodes a protein containing polyglutamine tracts and a leucine zipper. J Biol Chem. 1991, 266 (2): 863-872.
Wagner C, Blank M, Strohmann B, Schuller HJ: Overproduction of the Opi1 repressor inhibits transcriptional activation of structural genes required for phospholipid biosynthesis in the yeast Saccharomyces cerevisiae. Yeast. 1999, 15 (10A): 843-854. 10.1002/(SICI)1097-0061(199907)15:10A<843::AID-YEA424>3.0.CO;2-M.
Lo WS, Gamache ER, Henry KW, Yang D, Pillus L, Berger SL: Histone H3 phosphorylation can promote TBP recruitment through distinct promoter-specific mechanisms. EMBO J. 2005, 24 (5): 997-1008. 10.1038/sj.emboj.7600577.
Dietz M, Heyken WT, Hoppen J, Geburtig S, Schuller HJ: TFIIB and subunits of the SAGA complex are involved in transcriptional activation of phospholipid biosynthetic genes by the regulatory protein Ino2 in the yeast Saccharomyces cerevisiae. Mol Microbiol. 2003, 48 (4): 1119-1130. 10.1046/j.1365-2958.2003.03501.x.
Greenberg ML, Goldwasser P, Henry SA: Characterization of a yeast regulatory mutant constitutive for synthesis of inositol-1-phosphate synthase. Mol Gen Genet. 1982, 186 (2): 157-163. 10.1007/BF00331845.
Loewen CJ, Roy A, Levine TP: A conserved ER targeting motif in three families of lipid binding proteins and in Opi1p binds VAP. EMBO J. 2003, 22 (9): 2025-2035. 10.1093/emboj/cdg201.
Keleher CA, Redd MJ, Schultz J, Carlson M, Johnson AD: Ssn6-Tup1 is a general repressor of transcription in yeast. Cell. 1992, 68 (4): 709-719. 10.1016/0092-8674(92)90146-4.
Smith RL, Johnson AD: Turning genes off by Ssn6-Tup1: a conserved system of transcriptional repression in eukaryotes. Trends Biochem Sci. 2000, 25 (7): 325-330. 10.1016/S0968-0004(00)01592-9.
Wagner C, Dietz M, Wittmann J, Albrecht A, Schuller HJ: The negative regulator Opi1 of phospholipid biosynthesis in yeast contacts the pleiotropic repressor Sin3 and the transcriptional activator Ino2. Mol Microbiol. 2001, 41 (1): 155-166. 10.1046/j.1365-2958.2001.02495.x.
Wang H, Stillman DJ: Transcriptional repression in Saccharomyces cerevisiae by a SIN3-LexA fusion protein. Mol Cell Biol. 1993, 13 (3): 1805-1814.
Vidal M, Strich R, Esposito RE, Gaber RF: RPD1 (SIN3/UME4) is required for maximal activation and repression of diverse yeast genes. Mol Cell Biol. 1991, 11 (12): 6306-6316.
Wang H, Clark I, Nicholson PR, Herskowitz I, Stillman DJ: The Saccharomyces cerevisiae SIN3 gene, a negative regulator of HO, contains four paired amphipathic helix motifs. Mol Cell Biol. 1990, 10 (11): 5927-5936.
Kasten MM, Dorland S, Stillman DJ: A large protein complex containing the yeast Sin3p and Rpd3p transcriptional regulators. Mol Cell Biol. 1997, 17 (8): 4852-4858.
Heyken WT, Repenning A, Kumme J, Schuller HJ: Constitutive expression of yeast phospholipid biosynthetic genes by variants of Ino2 activator defective for interaction with Opi1 repressor. Mol Microbiol. 2005, 56 (3): 696-707. 10.1111/j.1365-2958.2004.04499.x.
Nikoloff DM, Henry SA: Genetic analysis of yeast phospholipid biosynthesis. Annu Rev Genet. 1991, 25: 559-583. 10.1146/annurev.ge.25.120191.003015.
Jesch SA, Zhao X, Wells MT, Henry SA: Genome-wide analysis reveals inositol, not choline, as the major effector of Ino2p-Ino4p and unfolded protein response target gene expression in yeast. J Biol Chem. 2005, 280 (10): 9106-9118. 10.1074/jbc.M411770200.
Lopes JM, Henry SA: Interaction of trans and cis regulatory elements in the INO1 promoter of Saccharomyces cerevisiae. Nucleic Acids Res. 1991, 19 (14): 3987-3994. 10.1093/nar/19.14.3987.
Bailis AM, Poole MA, Carman GM, Henry SA: The Membrane-Associated Enzyme Phosphatidylserine Synthase Is Regulated at the Level of Messenger-Rna Abundance. Mol Cell Biol. 1987, 7 (1): 167-176.
Hiesinger M, Wagner C, Schuller HJ: The acetyl-CoA synthetase gene ACS2 of the yeast Saccharomyces cerevisiae is coregulated with structural genes of fatty acid biosynthesis by the transcriptional activators Ino2p and Ino4p. FEBS Lett. 1997, 415 (1): 16-20. 10.1016/S0014-5793(97)01085-5.
Hasslacher M, Ivessa AS, Paltauf F, Kohlwein SD: Acetyl-CoA carboxylase from yeast is an essential enzyme and is regulated by factors that control phospholipid metabolism. J Biol Chem. 1993, 268 (15): 10946-10952.
Schwank S, Ebbert R, Rautenstrauss K, Schweizer E, Schuller HJ: Yeast transcriptional activator INO2 interacts as an Ino2p/Ino4p basic helix-loop-helix heteromeric complex with the inositol/choline-responsive element necessary for expression of phospholipid biosynthetic genes in Saccharomyces cerevisiae. Nucleic Acids Res. 1995, 23 (2): 230-237. 10.1093/nar/23.2.230.
Schuller HJ, Hahn A, Troster F, Schutz A, Schweizer E: Coordinate genetic control of yeast fatty acid synthase genes FAS1 and FAS2 by an upstream activation site common to genes involved in membrane lipid biosynthesis. EMBO J. 1992, 11 (1): 107-114.
Hoppen J, Repenning A, Albrecht A, Geburtig S, Schuller HJ: Comparative analysis of promoter regions containing binding sites of the heterodimeric transcription factor Ino2/Ino4 involved in yeast phospholipid biosynthesis. Yeast. 2005, 22 (8): 601-613. 10.1002/yea.1209.
Schuller HJ, Schorr R, Hoffmann B, Schweizer E: Regulatory gene INO4 of yeast phospholipid biosynthesis is positively autoregulated and functions as a transactivator of fatty acid synthase genes FAS1 and FAS2 from Saccharomyces cerevisiae. Nucleic Acids Res. 1992, 20 (22): 5955-5961. 10.1093/nar/20.22.5955.
Ambroziak J, Henry SA: INO2 and INO4 gene products, positive regulators of phospholipid biosynthesis in Saccharomyces cerevisiae, form a complex that binds to the INO1 promoter. J Biol Chem. 1994, 269 (21): 15344-15349.
Hoshizaki DK, Hill JE, Henry SA: The Saccharomyces cerevisiae INO4 gene encodes a small, highly basic protein required for derepression of phospholipid biosynthetic enzymes. J Biol Chem. 1990, 265 (8): 4736-4745.
Kumme J, Dietz M, Wagner C, Schuller HJ: Dimerization of yeast transcription factors Ino2 and Ino4 is regulated by precursors of phospholipid biosynthesis mediated by Opi1 repressor. Curr Genet. 2008, 54 (1): 35-45. 10.1007/s00294-008-0197-7.
Chen M, Lopes JM: Multiple basic helix-loop-helix proteins regulate expression of the ENO1 gene of Saccharomyces cerevisiae. Eukaryot Cell. 2007, 6 (5): 786-796. 10.1128/EC.00383-06.
Hosaka K, Nikawa J, Kodaki T, Yamashita S: Cloning and characterization of the SCS1 gene required for the expression of genes in yeast phospholipid synthesis. J Biochem. 1994, 115 (1): 131-136.
Schwank S, Hoffmann B, Sch-uller HJ: Influence of gene dosage and autoregulation of the regulatory genes INO2 and INO4 on inositol/choline-repressible gene transcription in the yeast Saccharomyces cerevisiae. Curr Genet. 1997, 31 (6): 462-468. 10.1007/s002940050231.
Loewen CJ, Gaspar ML, Jesch SA, Delon C, Ktistakis NT, Henry SA, Levine TP: Phospholipid metabolism regulated by a transcription factor sensing phosphatidic acid. Science. 2004, 304 (5677): 1644-1647. 10.1126/science.1096083.
Canelas AB, Harrison N, Fazio A, Zhang J, Pitkanen JP, van den Brink J, Bakker BM, Bogner L, Bouwman J, Castrillo JI: Integrated multilaboratory systems biology reveals differences in protein metabolism between two reference yeast strains. Nat Commun. 2010, 1: 145-10.1038/ncomms1150.
van Dijken JP, Bauer J, Brambilla L, Duboc P, Francois JM, Gancedo C, Giuseppin MLF, Heijnen JJ, Hoare M, Lange HC: An interlaboratory comparison of physiological and genetic properties of four Saccharomyces cerevisiae strains. Enzyme Microb Tech. 2000, 26 (9-10): 706-714. 10.1016/S0141-0229(00)00162-9.
Zhang J, Vaga S, Chumnanpuen P, Kumar R, Vemuri GN, Aebersold R, Nielsen J: Mapping the interaction of Snf1 with TORC1 in Saccharomyces cerevisiae. Mol Syst Biol. 2011, 7:
Chumnanpuen P, Zhang J, Nookaew I, Nielsen J: Integrated analysis of the transcriptome-lipidome reveals the co-influences of inositol-choline and Snf1 in controlling lipid biosynthesis in yeast. Molecular Genetics and Genomics: MGG. 2012, 287 (7): 541-554. 10.1007/s00438-012-0697-5.
Klig LS, Homann MJ, Carman GM, Henry SA: Coordinate Regulation of Phospholipid Biosynthesis in Saccharomyces-Cerevisiae - Pleiotropically Constitutive Opil Mutant. J Bacteriol. 1985, 162 (3): 1135-1141.
Zaldivar J, Borges A, Johansson B, Smits HP, Villas-Boas SG, Nielsen J, Olsson L: Fermentation performance and intracellular metabolite patterns in laboratory and industrial xylose-fermenting Saccharomyces cerevisiae. Appl Microbiol Biotechnol. 2002, 59 (4-5): 436-442. 10.1007/s00253-002-1056-y.
Bligh EG, Dyer WJ: A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1959, 37 (8): 911-917. 10.1139/o59-099.
Khoomrung S, Chumnanpuen P, Jansa-Ard S, Stahlman M, Nookaew I, Boren J, Nielsen J: Rapid quantification of yeast lipid using microwave-assisted total lipid extraction and HPLC-CAD. Analytical chemistry. 2013, 85 (10): 4912-4919. 10.1021/ac3032405.
Khoomrung S, Chumnanpuen P, Jansa-ard S, Nookaew I, Nielsen J: Fast and accurate preparation fatty acid methyl esters by microwave-assisted derivatization in yeast Saccharomyces cerevisiae. Applied Microbiology and Biotechnology. 2012, 94 (6): 1637-1646. 10.1007/s00253-012-4125-x.
Nookaew I, Jewett MC, Meechai A, Thammarongtham C, Laoteng K, Cheevadhanarak S, Nielsen J, Bhumiratana S: The genome-scale metabolic model iIN800 of Saccharomyces cerevisiae and its validation: a scaffold to query lipid metabolism. BMC Syst Biol. 2008, 2: 71-10.1186/1752-0509-2-71.
Oliveira AP, Patil KR, Nielsen J: Architecture of transcriptional regulatory circuits is knitted over the topology of bio-molecular interaction networks. BMC Syst Biol. 2008, 2: 17-10.1186/1752-0509-2-17.
Varemo L, Nielsen J, Nookaew I: Enriching the gene set analysis of genome-wide data by incorporating directionality of gene expression and combining statistical hypotheses and methods. Nucleic Acids Research. 2013, 41 (8): 4378-4391. 10.1093/nar/gkt111.
Graves JA, Henry SA: Regulation of the yeast INO1 gene. The products of the INO2, INO4 and OPI1 regulatory genes are not required for repression in response to inositol. Genetics. 2000, 154 (4): 1485-1495.
Tu BP, Kudlicki A, Rowicka M, McKnight SL: Logic of the yeast metabolic cycle: temporal compartmentalization of cellular processes. Science. 2005, 310 (5751): 1152-1158. 10.1126/science.1120499.
Chirala SS, Zhong Q, Huang W, al-Feel W: Analysis of FAS3/ACC regulatory region of Saccharomyces cerevisiae: identification of a functional UASINO and sequences responsible for fatty acid mediated repression. Nucleic Acids Res. 1994, 22 (3): 412-418. 10.1093/nar/22.3.412.
Schuller HJ, Richter K, Hoffmann B, Ebbert R, Schweizer E: DNA binding site of the yeast heteromeric Ino2p/Ino4p basic helix-loop-helix transcription factor: structural requirements as defined by saturation mutagenesis. FEBS Lett. 1995, 370 (1-2): 149-152. 10.1016/0014-5793(95)00818-T.
Tehlivets O: Homocysteine as a risk factor for atherosclerosis: is its conversion to s-adenosyl-L-homocysteine the key to deregulated lipid metabolism?. J Lipids. 2011, 2011: 702853-
Chen M, Hancock LC, Lopes JM: Transcriptional regulation of yeast phospholipid biosynthetic genes. Biochim Biophys Acta. 2007, 1771 (3): 310-321. 10.1016/j.bbalip.2006.05.017.
Lee TI, Rinaldi NJ, Robert F, Odom DT, Bar-Joseph Z, Gerber GK, Hannett NM, Harbison CT, Thompson CM, Simon I: Transcriptional regulatory networks in Saccharomyces cerevisiae. Science. 2002, 298 (5594): 799-804. 10.1126/science.1075090.
Ernst J, Vainas O, Harbison CT, Simon I, Bar-Joseph Z: Reconstructing dynamic regulatory maps. Mol Syst Biol. 2007, 3: 74-
Petiot A, Pattingre S, Arico S, Meley D, Codogno P: Diversity of signaling controls of macroautophagy in mammalian cells. Cell Struct Funct. 2002, 27 (6): 431-441. 10.1247/csf.27.431.
Maere S, Heymans K, Kuiper M: BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics. 2005, 21 (16): 3448-3449. 10.1093/bioinformatics/bti551.
Acknowledgements
We would like to thank Dr. Jie Zhang and Dr. Goutham Vemuri for kindly assistance with fermentation and microarray data acquisition, and Leif Varemo for kindly help with R script for transcriptome data analysis package.
Declarations
Part of this work was financed by Chalmers Foundation, the Knut and Alice Wallenberg Foundation and the Swedish Research Council (Vetenskapsrådet). We also acknowledge funding from the EU-funded project UNICELLSYS.
This article has been published as part of BMC Systems Biology Volume 7 Supplement 2, 2013: Twelfth International Conference on Bioinformatics (InCoB2013): Bioinformatics. The full contents of the supplement are available online at http://www.biomedcentral.com/bmcsystbiol/supplements/7/S3.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
PC conducted all the experiments, analyzed the data and prepared the manuscript. IN assisted with the data analysis for transcriptome and lipid analysis. JN edited and approved manuscript.
Electronic supplementary material
12918_2013_1164_MOESM1_ESM.pdf
Additional file 1:Supplementary Information to "Integrated analysis of the transcriptome-lipidome reveals the effects of INO-level (INO2 and INO4) on lipid metabolism in yeast"(PDF 2 MB)
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly credited. The Creative Commons Public Domain Dedication waiver ( https://creativecommons.org/publicdomain/zero/1.0/ ) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Chumnanpuen, P., Nookaew, I. & Nielsen, J. Integrated analysis, transcriptome-lipidome, reveals the effects of INO- level (INO2 and INO4) on lipid metabolism in yeast. BMC Syst Biol 7 (Suppl 3), S7 (2013). https://doi.org/10.1186/1752-0509-7-S3-S7
Published:
DOI: https://doi.org/10.1186/1752-0509-7-S3-S7